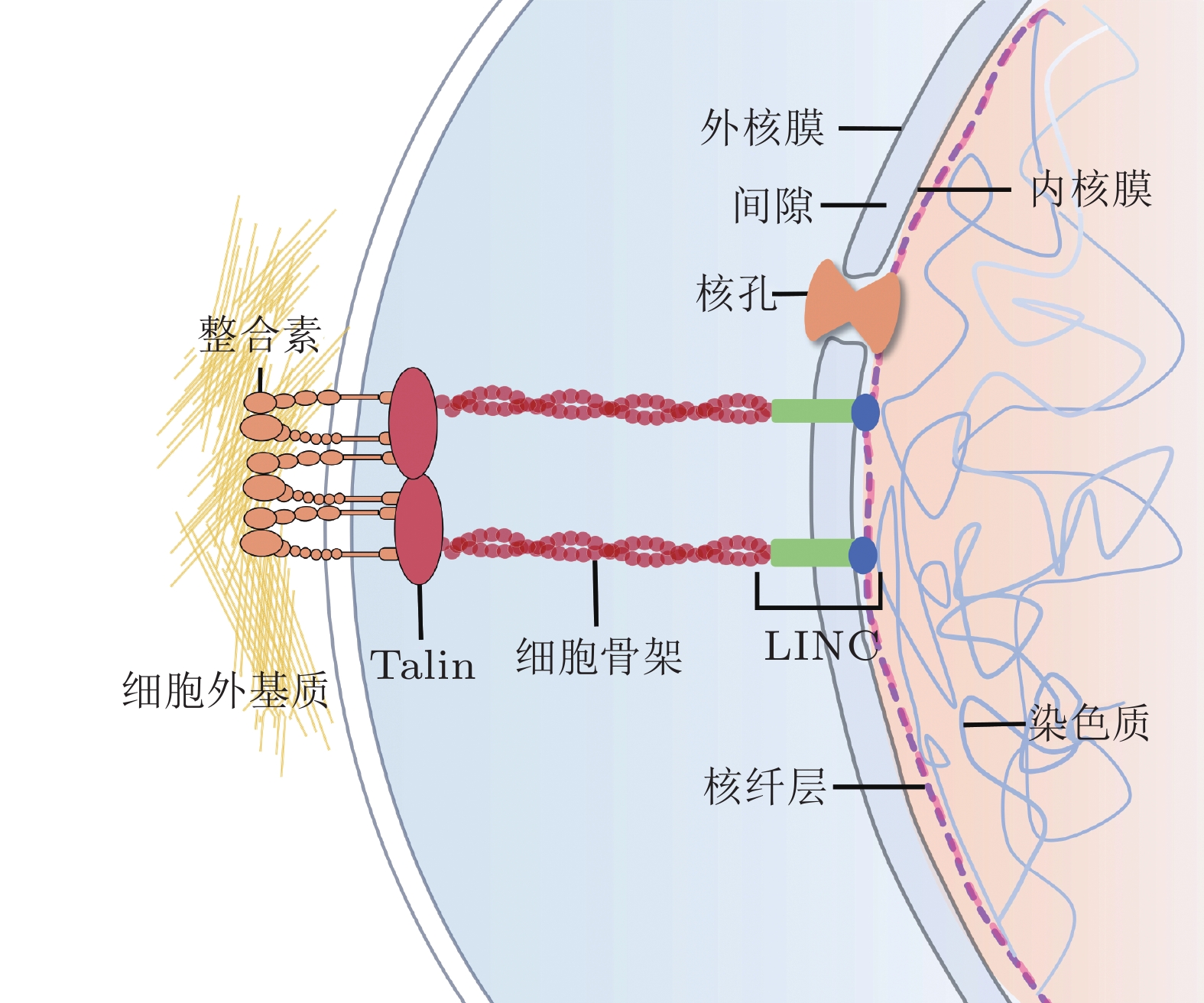
-
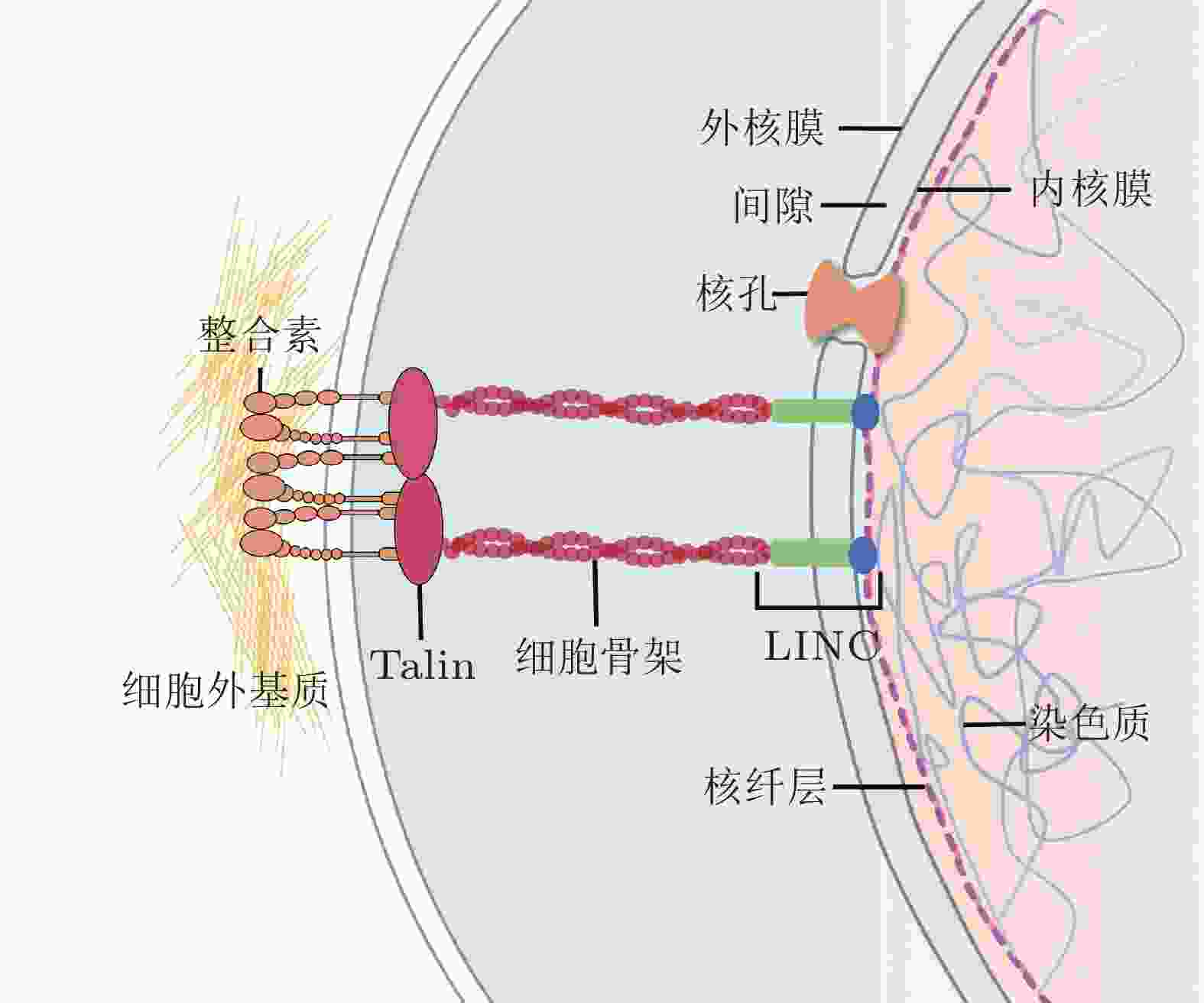
摘要: 细胞核是细胞内最大、刚度最高的细胞器, 也是遗传信息储存、复制和转录的中心场所, 在生命活动调节方面发挥关键作用. 研究表明, 生物力学因素在细胞核结构和功能的动态变化调控中至关重要. 核骨架蛋白和核孔复合体作为细胞核典型的力学响应组分, 在维持细胞核形态结构的同时将细胞骨架传递而来的力呈递至染色质, 进而影响染色质构象、基因表达等一系列细胞核相关活动, 调控细胞功能. 细胞核及其组分对应力信号的感知和转导已成为生物力学研究的前沿热点之一. 为更深入地认识生理和病理状态下细胞核的力学特性, 揭示其在细胞命运决定中的作用和机制, 本文总结了细胞核生物力学相关的研究进展, 重点介绍了细胞核骨架、核孔复合体和染色质的物理结构、力学响应过程、各组分的相互作用以及用于细胞核生物力学研究的技术进展, 最后总结了细胞核与早衰综合症、神经退行性疾病和心血管疾病的关系, 并对细胞核生物力学未来发展进行了展望.Abstract: The nucleus, which plays crucial roles in regulating life activities, is the largest and stiffest organelle in the cell and serves as the center of genetic information storage, replication, and transcription. Biomechanical factors have been shown to be of paramount importance in regulating dynamic changes of nuclear structures and functions. As typical mechanical responsive elements of the nucleus, nucleoskeletal proteins and nuclear pore complexes maintain the morphology and structure of the nucleus, and transmit mechanical forces from the cytoskeleton to chromatin. These factors affect a range of nuclear-related processes, including chromatin conformation and gene expression, and ultimately regulate cellular functions. The sensation and transduction of mechanical signals via nuclear components are one of the emerging cutting edges in biomechanics. To gain a deeper understanding of the mechanical properties of the nucleus in physiological and pathological states, and to elucidate its roles and mechanisms in cell fate determination, this review summarizes the research progresses related to nuclear biomechanics, focusing on the physical structures of nucleoskeleton, nuclear pore complex, and chromatin, the processes of mechanical responses, the interactions among these nuclear components, and the technological advances in nuclear biomechanics. Finally, the relationship between nucleus and the progeria, neurodegeneration or cardiovascular diseases and the future advancements of nuclear biomechanics are prospected.
-
Key words:
- nuclear biomechanics /
- mechanotransduction /
- nucleoskeleton /
- nuclear pore complex /
- chromatin
-
表 1 不同骨架蛋白之间的力学特性比较
蛋白名称 弹性模量 (dyn/cm2) 应变硬化 (%) 韧性 (%) 工程应变 (%) 相角δ (°) F-actin ~ 10
(Xu et al. 2000)20
(Janmey et al. 1991)10
(Xu et al. 2000)- 30
(Xu et al. 2000)Keratin 5-7
(Yamada et al. 2003)100
(Yamada et al. 2003)100-200
(Yamada et al. 2003)45
(Guthold et al. 2007)4-5
(Yamada et al. 2003)Vimentin 4
(Panorchan et al. 2004)- 12
(Panorchan et al. 2004)205
(Qin et al. 2009)10
(Panorchan et al. 2004)Lamin 1
(Panorchan et al. 2004)200
(Panorchan et al. 2004)200
(Panorchan et al. 2004)250
(Sapra et al. 2020)10
(Panorchan et al. 2004)表 2 使用不同技术测量不同类型细胞核刚度的实例
细胞类型 刚度 (kPa) 检测技术 参考文献 小鼠胚胎成纤维细胞 0.21 ± 0.01 微吸管 (Rowat et al. 2006) 人宫颈癌细胞
(Hela)0.41 ± 0.08 微吸管 (Rowat et al. 2005) 牛胸主动脉内皮细胞 0.42 ± 0.12 微吸管 (Deguchi et al. 2005) 大鼠肺上皮细胞 3.10 ± 1.50 AFM (Azeloglu et al. 2008) 大鼠肺成纤维细胞 3.30 ± 0.80 AFM (Azeloglu et al. 2008) 人瓣膜间质细胞 9.29 ± 1.80 AFM (Liu H et al. 2014) 人脐静脉内皮细胞 7.22 ± 0.46 AFM (Mathur et al. 2000) 人宫颈癌细胞
(Hela)6.70 ± 2.20 AFM (Nagayama et al. 2015) 人血管平滑肌细胞 23.20 ± 14.30 AFM (Nagayama et al. 2015) 人巩膜静脉窦内皮细胞 1.19 ± 0.19 AFM (Vargas-Pinto et al. 2013) 人髓系白血病细胞 0.13 ± 0.06 光镊 (Zhou Z L et al. 2014) 人肌肉上皮细胞 0.02 ± 0.01 光镊 (Coceano et al. 2016) 人乳腺癌细胞
(MCF-7)0.03 ± 0.02 光镊 (Coceano et al. 2016) 人乳腺癌细胞
(MDA-MB-231)0.02 ± 0.01 光镊 (Coceano et al. 2016) -
[1] 吕东媛, 周吕文, 龙勉. 2017. 干细胞的生物力学研究. 力学进展, 47: 534-585 doi: 10.6052/1000-0992-16-041 [2] 张欢, 赵国清, 冯锦腾, 2023. 力敏感受体介导细胞功能调控的力学生物学研究. 力学进展, 53 : 48-153 [3] Abels J A, Moreno-Herrero F, van der Heijden T, et al. 2005. Single-molecule measurements of the persistence length of double-stranded RNA. Biophysical Journal, 88: 2737-2744. doi: 10.1529/biophysj.104.052811 [4] Agbleke A A, Amitai A, Buenrostro J D, et al. 2020. Advances in chromatin and chromosome research: Perspectives from multiple fields. Mol Cell, 79: 881-901. doi: 10.1016/j.molcel.2020.07.003 [5] Alhmoud H, Alkhaled M, Kaynak B E, et al. 2023. Leveraging the elastic deformability of polydimethylsiloxane microfluidic channels for efficient intracellular delivery. Lab Chip, 23: 714-726. doi: 10.1039/D2LC00692H [6] Alisafaei F, Jokhun D S, Shivashankar G V, et al. 2019. Regulation of nuclear architecture, mechanics, and nucleocytoplasmic shuttling of epigenetic factors by cell geometric constraints. Proc Natl Acad Sci U S A, 116: 13200-13209. doi: 10.1073/pnas.1902035116 [7] Amemiya S. 2021. Nanoelectrochemical study of molecular transport through the nuclear pore complex. Chem Rec, 21 : 1430-1441. [8] Antonacci G, de Turris V, Rosa A, et al. 2018. Background-deflection Brillouin microscopy reveals altered biomechanics of intracellular stress granules by ALS protein FUS. Commun Biol, 1: 139. doi: 10.1038/s42003-018-0148-x [9] Ashkin A. 1970. Acceleration and trapping of particles by radiation pressure. Physical Review Letters, 24 : 156-159. [10] Ashkin A, Dziedzic J M, Bjorkholm J E, et al. 1986. Observation of a single-beam gradient force optical trap for dielectric particles. Optics Letters, 11: 288-290. doi: 10.1364/OL.11.000288 [11] Ashkin A, Dziedzic J M, Yamane T. 1987. Optical trapping and manipulation of single cells using infrared laser beams. Nature, 330 : 769-771. [12] Azeloglu E U, Bhattacharya J, Costa K D. 2008. Atomic force microscope elastography reveals phenotypic differences in alveolar cell stiffness. Journal of Applied Physiology, 105 : 652-661. [13] Baarlink C, Wang H, Grosse R. 2013. Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science, 340 : 864-867. [14] Ballatore C, Lee V M, Trojanowski J Q. 2007. Tau-mediated neurodegeneration in Alzheimer's disease and related disorders. Nat Rev Neurosci, 8 : 663-672. [15] Balzarotti F, Eilers Y, Gwosch K C, et al. 2017. Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes. Science, 355: 606-612. doi: 10.1126/science.aak9913 [16] Batha L, Aziz M A, Zhra M, et al. 2023. Differential gene expression signatures and cellular Signaling pathways induced by Lamin A/C transcript variants in MCF7 cell line. Front Biosci (Landmark Ed), 28: 113. doi: 10.31083/j.fbl2806113 [17] Beck M, Hurt E. 2017. The nuclear pore complex: understanding its function through structural insight. Nature Reviews Molecular Cell Biology, 18 : 73-89. [18] Belin B J, Lee T, Mullins R D. 2015. DNA damage induces nuclear actin filament assembly by Formin -2 and Spire-½ that promotes efficient DNA repair. [corrected]. Elife, 4: e07735. [19] Binnig G, Quate C F, Gerber C. 1986. Atomic force microscope. Physical Review Letters, 56: 930-933. [20] Bintu B, Mateo L J, Su J-H, et al. 2018. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science, 362: eaau1783. doi: 10.1126/science.aau1783 [21] Bley C J, Nie S, Mobbs G W, et al. 2022. Architecture of the cytoplasmic face of the nuclear pore. Science, 376: eabm9129. doi: 10.1126/science.abm9129 [22] Blobel G. 1985. Gene gating: a hypothesis. Proceedings of the National Academy of Sciences, 82: 8527-8529. [23] Bloom G S. 2014. Amyloid-β and tau: the trigger and bullet in Alzheimer disease pathogenesis. JAMA Neurol, 71 : 505-508. [24] Bohnsack M T, Stüven T, Kuhn C, et al. 2006. A selective block of nuclear actin export stabilizes the giant nuclei of Xenopus oocytes. Nat Cell Biol, 8: 257-263. doi: 10.1038/ncb1357 [25] Booth E A, Spagnol S T, Alcoser T A, et al. 2015. Nuclear stiffening and chromatin softening with progerin expression leads to an attenuated nuclear response to force. Soft Matter, 11: 6412-6418. doi: 10.1039/C5SM00521C [26] Bouwman B A M, Crosetto N, Bienko M. 2022. The era of 3D and spatial genomics. Trends Genet, 38: 1062-1075. [27] Boyle A P, Davis S, Shulha H P, et al. 2008. High-resolution mapping and characterization of open chromatin across the genome. Cell, 132: 311-322. doi: 10.1016/j.cell.2007.12.014 [28] Brickner D G, Cajigas I, Fondufe-Mittendorf Y, et al. 2007. H2A. Z-Mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLOS Biology, 5: e81. [29] Brillouin L. 1922. Diffusion de la lumière et des rayons X par un corps transparent homogène. Annales de Physique, 9 : 88-122. [30] Buchwalter A, Kaneshiro J M, Hetzer M W. 2019. Coaching from the sidelines: the nuclear periphery in genome regulation. Nat Rev Genet, 20 : 39-50. [31] Buenrostro J D, Giresi P G, Zaba L C, et al. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods, 10: 1213-1218. doi: 10.1038/nmeth.2688 [32] Burdine R D, Preston C C, Leonard R J, et al. 2020. Nucleoporins in cardiovascular disease. J Mol Cell Cardiol, 141: 43-52. doi: 10.1016/j.yjmcc.2020.02.010 [33] Byun S, Son S, Amodei D, et al. 2013. Characterizing deformability and surface friction of cancer cells. Proc Natl Acad Sci U S A, 110: 7580-7585. doi: 10.1073/pnas.1218806110 [34] Cain N E, Starr D A. 2015. Sun proteins and nuclear envelope spacing. Nucleus, 6 : 2-7. [35] Carley E, Stewart R M, Zieman A, et al. 2021. The LINC complex transmits integrin-dependent tension to the nuclear lamina and represses epidermal differentiation. Elife, 10. [36] Casolari J M, Brown C R, Komili S, et al. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell, 117: 427-439. doi: 10.1016/S0092-8674(04)00448-9 [37] Castano E, Philimonenko V V, Kahle M, et al. 2010. Actin complexes in the cell nucleus: new stones in an old field. Histochem Cell Biol, 133: 607-626. doi: 10.1007/s00418-010-0701-2 [38] Chaudhuri O, Gu L, Klumpers D, et al. 2016. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat Mater, 15: 326-334. doi: 10.1038/nmat4489 [39] Chen B, Gilbert L A, Cimini B A, et al. 2013. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell, 155: 1479-1491. doi: 10.1016/j.cell.2013.12.001 [40] Chen B C, Legant W R, Wang K, et al. 2014. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science, 346: 1257998. doi: 10.1126/science.1257998 [41] Cho S, Abbas A, Irianto J, et al. 2018. Progerin phosphorylation in interphase is lower and less mechanosensitive than lamin-A, C in iPS-derived mesenchymal stem cells. Nucleus, 9: 230-245. [42] Cho S, Vashisth M, Abbas A, et al. 2019. Mechanosensing by the lamina protects against nuclear rupture, DNA Damage, and Cell-Cycle Arrest. Dev Cell, 49 : 920-935. e925. [43] Christopher Wolf M R K M. 2009. Cellular mechanotransduction: Diverse perspectives from molecules to tissues[C]. Cambridge: Cambridge University Press. [44] Churnside A B, Sullan R M A, Nguyen D M, et al. 2012. Routine and timely sub-picoNewton force stability and precision for biological applications of atomic force microscopy. Nano Letters, 12: 3557-3561. doi: 10.1021/nl301166w [45] Cmarko D, Koberna K 2007. Electron microscopy in situ hybridization: tracking of DNA and RNA sequences at high resolution. Methods Mol Biol, 369 : 213-228. [46] Coceano G, Yousafzai M S, Ma W, et al. 2016. Investigation into local cell mechanics by atomic force microscopy mapping and optical tweezer vertical indentation. Nanotechnology, 27: 065102. doi: 10.1088/0957-4484/27/6/065102 [47] Coffinier C, Fong L G, Young S G 2010. LINCing lamin B2 to neuronal migration: Growing evidence for cell-specific roles of B-type lamins. Nucleus, 1 : 407-411. [48] Coffinier C, Jung H J, Nobumori C, et al. 2011. Deficiencies in lamin B1 and lamin B2 cause neurodevelopmental defects and distinct nuclear shape abnormalities in neurons. Mol Biol Cell, 22: 4683-4693. doi: 10.1091/mbc.e11-06-0504 [49] Combs C A. 2010. Fluorescence microscopy: a concise guide to current imaging methods. Curr Protoc Neurosci, Chapter 2: Unit2.1. [50] Coyne A N, Rothstein J D. 2022. Nuclear pore complexes — a doorway to neural injury in neurodegeneration. Nature Reviews Neurology, 18 : 348-362. [51] Cremer T, Cremer M. 2010. Chromosome territories. Cold Spring Harb Perspect Biol, 2 : a003889. [52] Crick F H C, Hughes A F W. 1950. The physical properties of cytoplasm: A study by means of the magnetic particle method part I. Experimental. Experimental Cell Research, 1 : 37-80. [53] D'Urso A, Takahashi Y H, Xiong B, et al. 2016. Set1/COMPASS and Mediator are repurposed to promote epigenetic transcriptional memory. Elife, 5. [54] Davey C A, Sargent D F, Luger K, et al. 2002. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9Å resolution. Journal of Molecular Biology, 319 : 1097-1113. [55] De Silva S, Fan Z, Kang B, et al. 2023. Nesprin-1: novel regulator of striated muscle nuclear positioning and mechanotransduction. Biochem Soc Trans, 51: 1331-1345. doi: 10.1042/BST20221541 [56] De Vries A H, Krenn B E, van Driel R, et al. 2007. Direct observation of nanomechanical properties of chromatin in living cells. Nano Lett, 7: 1424-1427. doi: 10.1021/nl070603+ [57] Deguchi S, Maeda K, Ohashi T, et al. 2005. Flow-induced hardening of endothelial nucleus as an intracellular stress-bearing organelle. Journal of Biomechanics, 38: 1751-1759. doi: 10.1016/j.jbiomech.2005.06.003 [58] Deguchi T, Iwanski M K, Schentarra E M, et al. 2023. Direct observation of motor protein stepping in living cells using MINFLUX. Science, 379: 1010-1015. doi: 10.1126/science.ade2676 [59] Dekker M, Van der Giessen E, Onck P R 2023. Phase separation of intrinsically disordered FG-Nups is driven by highly dynamic FG motifs. Proc Natl Acad Sci U S A, 120 : e2221804120. [60] Dieppois G, Iglesias N, Stutz F. 2006. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol Cell Biol, 26 : 7858-7870. [61] Dittrich P S, Manz A. 2006. Lab-on-a-chip: Microfluidics in drug discovery. Nat Rev Drug Discov, 5 : 210-218. [62] Doll S G, Meshkin H, Bryer A J, et al. 2022. Recognition of the TDP-43 nuclear localization signal by importin α1/β. Cell Rep, 39: 111007. doi: 10.1016/j.celrep.2022.111007 [63] Donnaloja F, Jacchetti E, Soncini M, et al. 2019. Mechanosensing at the nuclear envelope by nuclear pore complex stretch activation and its effect in physiology and pathology. Frontiers in Physiology, 10. [64] Dopie J, Skarp K-P, Kaisa Rajakylä E, et al. 2012. Active maintenance of nuclear actin by importin 9 supports transcription. Proceedings of the National Academy of Sciences, 109: E544-E552. [65] Downing T L, Soto J, Morez C, et al. 2013. Biophysical regulation of epigenetic state and cell reprogramming. Nat Mater, 12: 1154-1162. doi: 10.1038/nmat3777 [66] Earle A J, Kirby T J, Fedorchak G R, et al. 2020. Mutant lamins cause nuclear envelope rupture and DNA damage in skeletal muscle cells. Nat Mater, 19: 464-473. doi: 10.1038/s41563-019-0563-5 [67] Eastman T E, Zhu D-M. 1996. Adhesion forces between surface-modified AFM tips and a mica surface. Langmuir, 12 : 2859-2862. [68] Eftekharzadeh B, Daigle J G, Kapinos L E, et al. 2018. Tau protein disrupts nucleocytoplasmic transport in alzheimer's disease. Neuron, 99 : 925-940.e927. [69] Elosegui-Artola A, Andreu I, Beedle A E M, et al. 2017. Force triggers YAP nuclear entry by regulating transport across nuclear pores. Cell, 171 : 1397-1410. e1314. [70] Eltsov M, Sosnovski S, Olins A L, et al. 2014. ELCS in ice: Cryo-electron microscopy of nuclear envelope-limited chromatin sheets. Chromosoma, 123: 303-312. doi: 10.1007/s00412-014-0454-0 [71] Engler A J, Sen S, Sweeney H L, et al. 2006. Matrix elasticity directs stem cell lineage specification. Cell, 126: 677-689. doi: 10.1016/j.cell.2006.06.044 [72] Fan W, Adebowale K, Váncza L, et al. 2024. Matrix viscoelasticity promotes liver cancer progression in the pre-cirrhotic liver. Nature, 626: 635-642. doi: 10.1038/s41586-023-06991-9 [73] Fan X, Zhu L, Wang K, et al. 2017. Stiffness-controlled thermoresponsive hydrogels for cell harvesting with sustained mechanical memory. Advanced Healthcare Materials, 6: 1601152. doi: 10.1002/adhm.201601152 [74] Fernandez-Martinez J, Rout M P 2021. One ring to rule them all? Structural and functional diversity in the nuclear pore complex. Trends Biochem Sci, 46 : 595-607. [75] Fischer E G. 2020. Nuclear morphology and the biology of cancer cells. Acta Cytol, 64 : 511-519. [76] Fischer T, Hayn A, Mierke C T. 2020. Effect of nuclear stiffness on cell mechanics and migration of human breast cancer cells. Frontiers in Cell and Developmental Biology, 8. [77] Fisher P D E, Shen Q, Akpinar B, et al. 2018. A programmable DNA origami platform for organizing intrinsically disordered nucleoporins within nanopore confinement. ACS Nano, 12: 1508-1518. doi: 10.1021/acsnano.7b08044 [78] Fortuny A, Chansard A, Caron P, et al. 2021. Imaging the response to DNA damage in heterochromatin domains reveals core principles of heterochromatin maintenance. Nat Commun, 12: 2428. doi: 10.1038/s41467-021-22575-5 [79] Fukui K. 2016. Contribution of nanotechnology to chromosome science. Chromosome Science, 19 : 51-56. [80] Furusawa T, Rochman M, Taher L, et al. 2015. Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nat Commun, 6: 6138. doi: 10.1038/ncomms7138 [81] García-González A, Jacchetti E, Marotta R, et al. 2018. The effect of cell morphology on the permeability of the nuclear envelope to diffusive factors. Front Physiol, 9: 925. doi: 10.3389/fphys.2018.00925 [82] Gesson K, Rescheneder P, Skoruppa M P, et al. 2016. A-type lamins bind both hetero- and euchromatin, the latter being regulated by lamina-associated polypeptide 2 alpha. Genome Res, 26: 462-473. doi: 10.1101/gr.196220.115 [83] Gibson B A, Doolittle L K, Schneider M W G, et al. 2019. Organization of chromatin by intrinsic and regulated phase separation. Cell, 179 : 470-484. e421. [84] Golloshi R, Playter C, Freeman T F, et al. 2022. Constricted migration is associated with stable 3D genome structure differences in cancer cells. EMBO reports, 23: e52149. doi: 10.15252/embr.202052149 [85] Gu B, Comerci C J, McCarthy D G, et al. 2020. Opposing effects of cohesin and transcription on CTCF organization revealed by super-resolution imaging. Mol Cell, 80 : 699-711. e697. [86] Guilluy C, Osborne L D, Van Landeghem L, et al. 2014. Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nature Cell Biology, 16: 376-381. doi: 10.1038/ncb2927 [87] Gurusaran M, Davies O R 2021. A molecular mechanism for LINC complex branching by structurally diverse SUN-KASH 6: 6 assemblies. Elife, 10. [88] Gustafsson M G. 2005. Nonlinear structured-illumination microscopy: Wide-field fluorescence imaging with theoretically unlimited resolution. Proc Natl Acad Sci U S A, 102 : 13081-13086. [89] Guthold M, Liu W, Sparks E A, et al. 2007. A comparison of the mechanical and structural properties of fibrin fibers with other protein fibers. Cell Biochem Biophys, 49: 165-181. doi: 10.1007/s12013-007-9001-4 [90] Hagan I, Yanagida M. 1995. The product of the spindle formation gene sad1 + associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol, 129 : 1033-1047. [91] Hansen C L, Skordalakes E, Berger J M, et al. 2002. A robust and scalable microfluidic metering method that allows protein crystal growth by free interface diffusion. Proc Natl Acad Sci U S A, 99: 16531-16536. doi: 10.1073/pnas.262485199 [92] Hao Y, Cheng S, Tanaka Y, et al. 2020. Mechanical properties of single cells: Measurement methods and applications. Biotechnology Advances, 45: 107648. doi: 10.1016/j.biotechadv.2020.107648 [93] Hase M E, Kuznetsov N V, Cordes V C. 2001. Amino acid substitutions of coiled-coil protein tpr abrogate anchorage to the nuclear pore complex but not parallel, in-register homodimerization. Molecular Biology of the Cell, 12 : 2433-2452. [94] Hell S W. 2015. Nanoscopy with focused light (nobel lecture). Angew Chem Int Ed Engl, 54 : 8054-8066. [95] Hell S W, Wichmann J. 1994. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett, 19 : 780-782. [96] Hochmuth R M. 2000. Micropipette aspiration of living cells. Journal of Biomechanics, 33 : 15-22. [97] Hofmann W A, Stojiljkovic L, Fuchsova B, et al. 2004. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nature Cell Biology, 6: 1094-1101. doi: 10.1038/ncb1182 [98] Holaska J M. 2008. Emerin and the nuclear lamina in muscle and cardiac disease. Circ Res, 103 : 16-23. [99] Hoskins V E, Smith K, Reddy K L. 2021. The shifting shape of genomes: dynamics of heterochromatin interactions at the nuclear lamina. Curr Opin Genet Dev, 67 : 163-173. [100] Hsia C-R, McAllister J, Hasan O, et al. 2022. Confined migration induces heterochromatin formation and alters chromatin accessibility. iScience, 25: 104978. doi: 10.1016/j.isci.2022.104978 [101] Ibarra A, Benner C, Tyagi S, et al. 2016. Nucleoporin-mediated regulation of cell identity genes. Genes Dev, 30: 2253-2258. doi: 10.1101/gad.287417.116 [102] Inaga S, Tanaka K, Ushiki T. 2007. Transmission and scanning electron microscopy of mammalian metaphase chromosomes[C]//S. INAGA, K. TANAKA, T. USHIKI: 93-104. 10.1201/9781420044928-9. [103] Jaasma M J, Jackson W M, Keaveny T M. 2006. Measurement and characterization of whole-cell mechanical behavior. Ann Biomed Eng, 34 : 748-758. [104] Jain N, Iyer K V, Kumar A, et al. 2013. Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc Natl Acad Sci U S A, 110: 11349-11354. doi: 10.1073/pnas.1300801110 [105] Janmey P A, Euteneuer U, Traub P, et al. 1991. Viscoelastic properties of vimentin compared with other filamentous biopolymer networks. Journal of Cell Biology, 113: 155-160. doi: 10.1083/jcb.113.1.155 [106] Janota C S, Calero-Cuenca F J, Costa J, et al. 2017. SnapShot: Nucleo-cytoskeletal interactions. Cell, 169 : 970-970. e971. [107] Janota C S, Calero-Cuenca F J, Gomes E R. 2020. The role of the cell nucleus in mechanotransduction. Curr Opin Cell Biol, 63 : 204-211. [108] Joseph J, Dasso M. 2008. The nucleoporin Nup358 associates with and regulates interphase microtubules. FEBS Lett, 582 : 190-196. [109] Junker J P, Ziegler F, Rief M. 2009. Ligand-dependent equilibrium fluctuations of single calmodulin molecules. Science, 323 : 633-637. [110] Kaczmarczyk A, Brouwer T B, Pham C, et al. 2018. Probing chromatin structure with magnetic tweezers. Methods Mol Biol, 1814: 297-323. [111] Kalukula Y, Stephens A D, Lammerding J, et al. 2022. Mechanics and functional consequences of nuclear deformations. Nat Rev Mol Cell Biol, 23: 583-602. [112] Kaneshiro J M, Capitanio J S, Hetzer M W. 2023. Lamin B1 overexpression alters chromatin organization and gene expression. Nucleus, 14 : 2202548. [113] Kaul A, Bhattacharyya S, Ay F. 2020. Identifying statistically significant chromatin contacts from Hi-C data with FitHiC2. Nat Protoc, 15 : 991-1012. [114] Kawaguchi K, Ishiwata S I. 2001. Nucleotide-dependent single-to double-headed binding of kinesin. Science, 291 : 667-669. [115] Kechagia Z, Roca-Cusachs P. 2023. Cytoskeletal safeguards: Protecting the nucleus from mechanical perturbations. Current Opinion in Biomedical Engineering, 28 : 100494. [116] Keizer V I P, Grosse-Holz S, Woringer M, et al. 2022. Live-cell micromanipulation of a genomic locus reveals interphase chromatin mechanics. Science, 377: 489-495. doi: 10.1126/science.abi9810 [117] Kelpsch D J, Tootle T L. 2018. Nuclear actin: From discovery to function. The Anatomical Record, 301 : 1999-2013. [118] Khanna N, Zhang Y, Lucas J S, et al. 2019. Chromosome dynamics near the sol-gel phase transition dictate the timing of remote genomic interactions. Nat Commun, 10: 2771. doi: 10.1038/s41467-019-10628-9 [119] Killaars A R, Grim J C, Walker C J, et al. 2019. Extended exposure to stiff microenvironments leads to persistent chromatin remodeling in human mesenchymal stem cells. Advanced Science, 6: 1801483. doi: 10.1002/advs.201801483 [120] Killaars A R, Walker C J, Anseth K S. 2020. Nuclear mechanosensing controls MSC osteogenic potential through HDAC epigenetic remodeling. Proceedings of the National Academy of Sciences, 117 : 21258-21266. [121] Kmonickova V, Frolikova M, Steger K, et al. 2020. The role of the LINC complex in sperm development and function. International Journal of Molecular Sciences, 21: 9058. doi: 10.3390/ijms21239058 [122] Koski K J, Yarger J L. 2005. Brillouin imaging. Applied Physics Letters, 87. [123] Koster D A, Crut A, Shuman S, et al. 2010. Cellular strategies for regulating DNA supercoiling: A single-molecule perspective. Cell, 142: 519-530. doi: 10.1016/j.cell.2010.08.001 [124] Koster D A, Palle K, Bot E S M, et al. 2007. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature, 448: 213-217. doi: 10.1038/nature05938 [125] Krause M, te Riet J, Wolf K. 2013. Probing the compressibility of tumor cell nuclei by combined atomic force–confocal microscopy. Physical Biology, 10 : 065002. [126] Kruithof M, Chien F-T, Routh A, et al. 2009. Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30-nm chromatin fiber. Nature Structural & Molecular Biology, 16: 534-540. [127] Krull S, Thyberg J, Björkroth B, et al. 2004. Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Molecular Biology of the Cell, 15: 4261-4277. doi: 10.1091/mbc.e04-03-0165 [128] Kuhn T M, Pascual-Garcia P, Gozalo A, et al. 2019. Chromatin targeting of nuclear pore proteins induces chromatin decondensation. J Cell Biol, 218: 2945-2961. doi: 10.1083/jcb.201807139 [129] LaJoie D, Turkmen A M, Mackay D R, et al. 2022. A role for Nup153 in nuclear assembly reveals differential requirements for targeting of nuclear envelope constituents. Mol Biol Cell, 33: ar117. doi: 10.1091/mbc.E22-05-0189 [130] Lane N J. 1969. Intranuclear fibrillar bodies in actinomycin D-treated oocytes. J Cell Biol, 40 : 286-291. [131] Lansky Z, Braun M, Lüdecke A, et al. 2015. Diffusible crosslinkers generate directed forces in microtubule networks. Cell, 160: 1159-1168. doi: 10.1016/j.cell.2015.01.051 [132] Le Dour C, Chatzifrangkeskou M, Macquart C, et al. 2022. Actin-microtubule cytoskeletal interplay mediated by MRTF-A/SRF signaling promotes dilated cardiomyopathy caused by LMNA mutations. Nat Commun, 13: 7886. doi: 10.1038/s41467-022-35639-x [133] Le H Q, Ghatak S, Yeung C Y, et al. 2016. Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat Cell Biol, 18: 864-875. doi: 10.1038/ncb3387 [134] Le T T, Gao X, Park S H, et al. 2019. Synergistic coordination of chromatin torsional mechanics and topoisomerase activity. Cell, 179 : 619-631. e615. [135] Lee H G, Ueda M, Miyamoto Y, et al. 2006. Aberrant localization of importin alpha1 in hippocampal neurons in Alzheimer disease. Brain Res, 1124: 1-4. doi: 10.1016/j.brainres.2006.09.084 [136] Leone L, Colussi C, Gironi K, et al. 2019. Altered Nup153 expression impairs the function of cultured hippocampal neural stem cells isolated from a mouse model of alzheimer's disease. Mol Neurobiol, 56: 5934-5949. doi: 10.1007/s12035-018-1466-1 [137] Li C X, Talele N P, Boo S, et al. 2017. MicroRNA-21 preserves the fibrotic mechanical memory of mesenchymal stem cells. Nat Mater, 16: 379-389. doi: 10.1038/nmat4780 [138] Li P, Noegel A A. 2015. Inner nuclear envelope protein SUN1 plays a prominent role in mammalian mRNA export. Nucleic Acids Res, 43 : 9874-9888. [139] Light W H, Freaney J, Sood V, et al. 2013. A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol, 11: e1001524. doi: 10.1371/journal.pbio.1001524 [140] Liu H, Wen J, Xiao Y, et al. 2014. In situ mechanical characterization of the cell nucleus by atomic force microscopy. ACS Nano, 8: 3821-3828. doi: 10.1021/nn500553z [141] Liu J, Hetzer M W. 2022. Nuclear pore complex maintenance and implications for age-related diseases. Trends Cell Biol, 32 : 216-227. [142] Liu S, Yang H, Lu T J, et al. 2019. Electrostatic switching of nuclear basket conformations provides a potential mechanism for nuclear mechanotransduction. Journal of the Mechanics and Physics of Solids, 133: 103705. doi: 10.1016/j.jmps.2019.103705 [143] Liu X, Zhang Y, Chen Y, et al. 2017. In situ capture of chromatin interactions by biotinylated dCas9. Cell, 170 : 1028-1043. e1019. [144] Liu Y, Lu Y, Yang X, et al. 2017. Amplified stimulated emission in upconversion nanoparticles for super-resolution nanoscopy. Nature, 543: 229-233. doi: 10.1038/nature21366 [145] Liu Z, Mao S, Hu Y, et al. 2023. Hydrogel platform facilitating astrocytic differentiation through cell mechanosensing and YAP-mediated transcription. Mater Today Bio, 22: 100735. doi: 10.1016/j.mtbio.2023.100735 [146] Liu Z, Yan M, Liang Y, et al. 2019. Nucleoporin Seh1 interacts with Olig2/Brd7 to promote oligodendrocyte differentiation and myelination. Neuron, 102 : 587-601. e587. [147] Lu J, Wu T, Zhang B, et al. 2021. Types of nuclear localization signals and mechanisms of protein import into the nucleus. Cell Commun Signal, 19: 60. doi: 10.1186/s12964-021-00741-y [148] Lund E G, Duband-Goulet I, Oldenburg A, et al. 2015. Distinct features of lamin A-interacting chromatin domains mapped by ChIP-sequencing from sonicated or micrococcal nuclease-digested chromatin. Nucleus, 6: 30-39. doi: 10.4161/19491034.2014.990855 [149] Luo M, Yukawa H, Baba Y. 2022. Micro-/nano-fluidic devices and in vivo fluorescence imaging based on quantum dots for cytologic diagnosis. Lab on a Chip, 22 : 2223-2236. [150] Ma S, Zhang Y. 2020. Profiling chromatin regulatory landscape: insights into the development of ChIP-seq and ATAC-seq. Mol Biomed, 1 : 9. [151] Maier B, Bensimon D, Croquette V. 2000. Replication by a single DNA polymerase of a stretched single-stranded DNA. Proceedings of the National Academy of Sciences, 97 : 12002-12007. [152] Manosas M, Xi X G, Bensimon D, et al. 2010. Active and passive mechanisms of helicases. Nucleic Acids Research, 38: 5518-5526. doi: 10.1093/nar/gkq273 [153] Manz A, Harrison D J, Verpoorte E M J, et al. 1992. Planar chips technology for miniaturization and integration of separation techniques into monitoring systems: Capillary electrophoresis on a chip. Journal of Chromatography A, 593: 253-258. doi: 10.1016/0021-9673(92)80293-4 [154] Marco S, Neilson M, Moore M, et al. 2021. Nuclear-capture of endosomes depletes nuclear G-actin to promote SRF/MRTF activation and cancer cell invasion. Nat Commun, 12: 6829. doi: 10.1038/s41467-021-26839-y [155] Mardaryev A N, Gdula M R, Yarker J L, et al. 2014. P63 and Brg1 control developmentally regulated higher-order chromatin remodelling at the epidermal differentiation complex locus in epidermal progenitor cells. Development, 141: 101-111. doi: 10.1242/dev.103200 [156] Marti O, Drake B, Hansma P K. 1987. Atomic force microscopy of liquid‐covered surfaces: Atomic resolution images. Applied Physics Letters, 51 : 484-486. [157] Mathies L D, Lindsay J H, Handal A P, et al. 2020. SWI/SNF complexes act through CBP-1 histone acetyltransferase to regulate acute functional tolerance to alcohol. BMC Genomics, 21: 646. doi: 10.1186/s12864-020-07059-y [158] Mathur A B, Truskey G A, Monty Reichert W. 2000. Atomic force and total internal reflection fluorescence microscopy for the study of force transmission in endothelial cells. Biophysical Journal, 78 : 1725-1735. [159] Matković J, Ghosh S, Ćosić M, et al. 2022. Kinetochore- and chromosome-driven transition of microtubules into bundles promotes spindle assembly. Nat Commun, 13: 7307. doi: 10.1038/s41467-022-34957-4 [160] Matsuda A, Mofrad M R K. 2022. On the nuclear pore complex and its emerging role in cellular mechanotransduction. APL Bioengineering, 6. [161] McCauley M J, Huo R, Becker N, et al. 2019. Single and double box HMGB proteins differentially destabilize nucleosomes. Nucleic Acids Res, 47: 666-678. doi: 10.1093/nar/gky1119 [162] McDonald J C, Duffy D C, Anderson J R, et al. 2000. Fabrication of microfluidic systems in poly(dimethylsiloxane). Electrophoresis, 21: 27-40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C [163] McNamara L E, Burchmore R, Riehle M O, et al. 2012. The role of microtopography in cellular mechanotransduction. Biomaterials, 33: 2835-2847. doi: 10.1016/j.biomaterials.2011.11.047 [164] Meijering A E C, Sarlós K, Nielsen C F, et al. 2022. Nonlinear mechanics of human mitotic chromosomes. Nature, 605: 545-550. doi: 10.1038/s41586-022-04666-5 [165] Melters D P, Pitman M, Rakshit T, et al. 2019. Intrinsic elasticity of nucleosomes is encoded by histone variants and calibrated by their binding partners. Proc Natl Acad Sci U S A, 116: 24066-24074. doi: 10.1073/pnas.1911880116 [166] Meqbel B R M, Gomes M, Omer A, et al. 2022. LINCing senescence and nuclear envelope changes. Cells, 11. [167] Miron E, Oldenkamp R, Brown J M, et al. 2020. Chromatin arranges in chains of mesoscale domains with nanoscale functional topography independent of cohesin. Sci Adv, 6. [168] Morez C, Noseda M, Paiva M A, et al. 2015. Enhanced efficiency of genetic programming toward cardiomyocyte creation through topographical cues. Biomaterials, 70: 94-104. doi: 10.1016/j.biomaterials.2015.07.063 [169] Muchir A, Worman H J. 2019. Emery-Dreifuss muscular dystrophy: focal point nuclear envelope. Curr Opin Neurol, 32 : 728-734. [170] Mitchison J M. 1954. The mechanical properties of the cell surface : I. The cell elastimeter. Journal of Experimental Biology, 31 : 443-460. [171] Müller D J, Dufrêne Y F. 2008. Atomic force microscopy as a multifunctional molecular toolbox in nanobiotechnology. Nature Nanotechnology, 3 : 261-269. [172] Mumbach M R, Satpathy A T, Boyle E A, et al. 2017. Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat Genet, 49: 1602-1612. doi: 10.1038/ng.3963 [173] Na S, Collin O, Chowdhury F, et al. 2008. Rapid signal transduction in living cells is a unique feature of mechanotransduction. Proceedings of the National Academy of Sciences, 105: 6626-6631. doi: 10.1073/pnas.0711704105 [174] Nagayama K, Hamaji Y, Sato Y, et al. 2015. Mechanical trapping of the nucleus on micropillared surfaces inhibits the proliferation of vascular smooth muscle cells but not cervical cancer HeLa cells. Journal of Biomechanics, 48: 1796-1803. doi: 10.1016/j.jbiomech.2015.05.004 [175] Nava M M, Miroshnikova Y A, Biggs L C, et al. 2020. Heterochromatin-driven nuclear softening protects the genome against mechanical stress-induced damage. Cell, 181 : 800-817. e822. [176] Neelam S, Chancellor T J, Li Y, et al. 2015. Direct force probe reveals the mechanics of nuclear homeostasis in the mammalian cell. Proceedings of the National Academy of Sciences, 112: 5720-5725. doi: 10.1073/pnas.1502111112 [177] Neuman K C, Nagy A. 2008. Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nat Methods, 5 : 491-505. [178] Nmezi B, Xu J, Fu R, et al. 2019. Concentric organization of A- and B-type lamins predicts their distinct roles in the spatial organization and stability of the nuclear lamina. Proc Natl Acad Sci U S A, 116: 4307-4315. doi: 10.1073/pnas.1810070116 [179] Nozaki T, Imai R, Tanbo M, et al. 2017. Dynamic organization of chromatin domains revealed by super-resolution live-cell imaging. Mol Cell, 67 : 282-293. e287. [180] Oosterheert W, Klink B U, Belyy A, et al. 2022. Structural basis of actin filament assembly and aging. Nature, 611: 374-379. doi: 10.1038/s41586-022-05241-8 [181] Ovsiannikova N L, Lavrushkina S V, Ivanova A V, et al. 2021. Lamin A as a determinant of mechanical properties of the cell nucleus in health and disease. Biochemistry (Mosc), 86: 1288-1300. doi: 10.1134/S0006297921100102 [182] Padiath Q S, Saigoh K, Schiffmann R, et al. 2006. Lamin B1 duplications cause autosomal dominant leukodystrophy. Nat Genet, 38: 1114-1123. doi: 10.1038/ng1872 [183] Pajerowski J D, Dahl K N, Zhong F L, et al. 2007. Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci U S A, 104: 15619-15624. doi: 10.1073/pnas.0702576104 [184] Panorchan P, Schafer B W, Wirtz D, et al. 2004. Nuclear envelope breakdown requires overcoming the mechanical integrity of the nuclear lamina*. Journal of Biological Chemistry, 279: 43462-43467. doi: 10.1074/jbc.M402474200 [185] Paonessa F, Evans L D, Solanki R, et al. 2019. Microtubules deform the nuclear membrane and disrupt nucleocytoplasmic transport in Tau-mediated frontotemporal dementia. Cell Rep, 26 : 582-593. e585. [186] Pascual-Garcia P, Jeong J, Capelson M. 2014. Nucleoporin Nup98 associates with Trx/MLL and NSL histone-modifying complexes and regulates Hox gene expression. Cell Rep, 9 : 433-442. [187] Percipalle P, Vartiainen M. 2019. Cytoskeletal proteins in the cell nucleus: a special nuclear actin perspective. Molecular Biology of the Cell, 30 : 1781-1785. [188] Petchakup C, Yang H, Gong L, et al. 2022. Microfluidic impedance-deformability cytometry for label-free single neutrophil mechanophenotyping. Small, 18: e2104822. doi: 10.1002/smll.202104822 [189] Petrovic S, Samanta D, Perriches T, et al. 2022. Architecture of the linker-scaffold in the nuclear pore. Science, 376: eabm9798. doi: 10.1126/science.abm9798 [190] Peyro M, Dickson A M, Mofrad M R K. 2021. Nucleoporins' exclusive amino acid sequence features regulate their transient interaction with and selectivity of cargo complexes in the nuclear pore. Mol Biol Cell, 32 : ar31. [191] Pollard T D, Cooper J A. 2009. Actin, a central player in cell shape and movement. Science, 326 : 1208-1212. [192] Qin Z, Kreplak L, Buehler M J. 2009. Hierarchical structure controls nanomechanical properties of vimentin intermediate filaments. PLoS One, 4 : e7294. [193] Rabineau M, Flick F, Mathieu E, et al. 2015. Cell guidance into quiescent state through chromatin remodeling induced by elastic modulus of substrate. Biomaterials, 37: 144-155. doi: 10.1016/j.biomaterials.2014.10.023 [194] Rand R P, Burton A C. 1964. Mechanical properties of the red cell membrane: I. Membrane stiffness and intracellular pressure. Biophysical Journal, 4 : 115-135. [195] Rao S S, Huntley M H, Durand N C, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell, 159: 1665-1680. doi: 10.1016/j.cell.2014.11.021 [196] Rashid F, Liu W, Wang Q, et al. 2023. Mechanomemory in protein diffusivity of chromatin and nucleoplasm after force cessation. Proceedings of the National Academy of Sciences, 120: e2221432120. doi: 10.1073/pnas.2221432120 [197] Remer I, Shaashoua R, Shemesh N, et al. 2020. High-sensitivity and high-specificity biomechanical imaging by stimulated Brillouin scattering microscopy. Nature Methods, 17: 913-916. doi: 10.1038/s41592-020-0882-0 [198] Roberts A B, Zhang J, Raj Singh V, et al. 2021. Tumor cell nuclei soften during transendothelial migration. Journal of Biomechanics, 121: 110400. doi: 10.1016/j.jbiomech.2021.110400 [199] Robinson P J J, Fairall L, Huynh V A T, et al. 2006. EM measurements define the dimensions of the “30-nm” chromatin fiber: Evidence for a compact, interdigitated structure. Proceedings of the National Academy of Sciences, 103: 6506-6511. doi: 10.1073/pnas.0601212103 [200] Rowat A C, Foster L J, Nielsen M M, et al. 2005. Characterization of the elastic properties of the nuclear envelope. Journal of The Royal Society Interface, 2: 63-69. doi: 10.1098/rsif.2004.0022 [201] Roman M, Wrobel T P, Paluszkiewicz C, et al. 2020. Comparison between high definition FT-IR, Raman and AFM-IR for subcellular chemical imaging of cholesteryl esters in prostate cancer cells. J Biophotonics, 13: e201960094 [202] Rowat A C, Lammerding J, Ipsen J H. 2006. Mechanical properties of the cell nucleus and the effect of emerin deficiency. Biophys J, 91 : 4649-4664. [203] Sapra K T, Qin Z, Dubrovsky-Gaupp A, et al. 2020. Nonlinear mechanics of lamin filaments and the meshwork topology build an emergent nuclear lamina. Nat Commun, 11: 6205. doi: 10.1038/s41467-020-20049-8 [204] Scarcelli G, Yun S H. 2007. Confocal Brillouin microscopy for three-dimensional mechanical imaging. Nat Photonics, 2 : 39-43. [205] Schermelleh L, Carlton P M, Haase S, et al. 2008. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science, 320: 1332-1336. doi: 10.1126/science.1156947 [206] Schmid M, Arib G, Laemmli C, et al. 2006. Nup-PI: The nucleopore-promoter interaction of genes in yeast. Molecular Cell, 21: 379-391. doi: 10.1016/j.molcel.2005.12.012 [207] Scholz B A, Sumida N, de Lima C D M, et al. 2019. WNT signaling and AHCTF1 promote oncogenic MYC expression through super-enhancer-mediated gene gating. Nature Genetics, 51: 1723-1731. doi: 10.1038/s41588-019-0535-3 [208] Schrank B R, Aparicio T, Li Y, et al. 2018. Nuclear ARP2/3 drives DNA break clustering for homology-directed repair. Nature, 559: 61-66. doi: 10.1038/s41586-018-0237-5 [209] Schreiner S M, Koo P K, Zhao Y, et al. 2015. The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat Commun, 6: 7159. doi: 10.1038/ncomms8159 [210] Scott A K, Casas E, Schneider S E, et al. 2023. Mechanical memory stored through epigenetic remodeling reduces cell therapeutic potential. Biophysical Journal, 122: 1428-1444. doi: 10.1016/j.bpj.2023.03.004 [211] Serebryannyy L A, Parilla M, Annibale P, et al. 2016. Persistent nuclear actin filaments inhibit transcription by RNA polymerase II. Journal of Cell Science, 129: 3412-3425. [212] Shah P, Hobson C M, Cheng S, et al. 2021a. Nuclear deformation causes DNA damage by increasing replication stress. Curr Biol, 31 : 753-765. e756. [213] Shah P P, Lv W, Rhoades J H, et al. 2021b. Pathogenic LMNA variants disrupt cardiac lamina-chromatin interactions and de-repress alternative fate genes. Cell Stem Cell, 28 : 938-954. e939. [214] Shao X, Li Q, Mogilner A, et al. 2015. Mechanical stimulation induces formin-dependent assembly of a perinuclear actin rim. Proceedings of the National Academy of Sciences, 112: E2595-E2601. [215] Shao X, Liu Z, Mao S, et al. 2022. Unraveling the Mechanobiology underlying traumatic brain injury with advanced technologies and biomaterials. Advanced Healthcare Materials, 11: 2200760. doi: 10.1002/adhm.202200760 [216] Shi Q, Zheng L, Na J, et al. 2022. Fluid shear stress promotes periodontal ligament cells proliferation via p38-AMOT-YAP. Cell Mol Life Sci, 79: 551. doi: 10.1007/s00018-022-04591-w [217] Singh I, Lele T P. 2022. Nuclear morphological abnormalities in cancer: A search for unifying mechanisms. Results Probl Cell Differ, 70 : 443-467. [218] Smeets D, Markaki Y, Schmid V J, et al. 2014. Three-dimensional super-resolution microscopy of the inactive X chromosome territory reveals a collapse of its active nuclear compartment harboring distinct Xist RNA foci. Epigenetics Chromatin, 7: 8. doi: 10.1186/1756-8935-7-8 [219] Smith S B, Cui Y, Bustamante C. 1996. Overstretching B-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science, 271 : 795-799. [220] Smith S B, Finzi L, Bustamante C. 1992. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science, 258 : 1122-1126. [221] Snyers L, Löhnert R, Weipoltshammer K, et al. 2022. Emerin prevents BAF-mediated aggregation of lamin A on chromosomes in telophase to allow nuclear membrane expansion and nuclear lamina formation. Mol Biol Cell, 33: ar137. doi: 10.1091/mbc.E22-01-0007 [222] Solovei I, Cavallo A, Schermelleh L, et al. 2002. Spatial preservation of nuclear chromatin architecture during three-dimensional fluorescence in situ hybridization (3D-FISH). Experimental Cell Research, 276: 10-23. doi: 10.1006/excr.2002.5513 [223] Song Y, Soto J, Li S. 2020. Mechanical regulation of histone modifications and cell plasticity. Curr Opin Solid State Mater Sci, 24. [224] Spead O, Zaepfel B L, Rothstein J D 2022. Nuclear pore dysfunction in neurodegeneration. Neurotherapeutics, 19: 1050-1060. [225] Starr D A, Fridolfsson H N. 2010. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annual Review of Cell and Developmental Biology, 26 : 421-444. [226] Stephens A D, Banigan E J, Adam S A, et al. 2017. Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol Biol Cell, 28: 1984-1996. doi: 10.1091/mbc.e16-09-0653 [227] Stephens A D, Liu P Z, Kandula V, et al. 2019. Physicochemical mechanotransduction alters nuclear shape and mechanics via heterochromatin formation. Mol Biol Cell, 30: 2320-2330. doi: 10.1091/mbc.E19-05-0286 [228] Streichfuss M, Erbs F, Uhrig K, et al. 2011. Measuring forces between two single actin filaments during bundle formation. Nano Letters, 11: 3676-3680. doi: 10.1021/nl201630y [229] Strickfaden H, Tolsma T O, Sharma A, et al. 2020. Condensed chromatin behaves like a solid on the mesoscale in vitro and in living cells. Cell, 183 : 1772-1784. e1713. [230] Stumme-Diers M P, Banerjee S, Hashemi M, et al. 2018. Nanoscale dynamics of centromere nucleosomes and the critical roles of CENP-A. Nucleic Acids Res, 46: 94-103. doi: 10.1093/nar/gkx933 [231] Sánchez-López A, Espinós-Estévez C, González-Gómez C, et al. 2021. Cardiovascular progerin suppression and lamin A restoration rescue hutchinson-gilford progeria syndrome. Circulation, 144: 1777-1794. doi: 10.1161/CIRCULATIONAHA.121.055313 [232] Sun J, Chen J, Mohagheghian E, et al. 2020. Force-induced gene up-regulation does not follow the weak power law but depends on H3K9 demethylation. Sci Adv, 6: eaay9095. doi: 10.1126/sciadv.aay9095 [233] Sun J, Shi Y, Yildirim E. 2019. The nuclear pore complex in cell type-specific chromatin structure and gene regulation. Trends Genet, 35 : 579-588. [234] Sung M H, Baek S, Hager G L. 2016. Genome-wide footprinting: ready for prime time? Nat Methods, 13 : 222-228. [235] Swift J, Ivanovska I L, Buxboim A, et al. 2013. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science, 341: 1240104. doi: 10.1126/science.1240104 [236] Tajik A, Zhang Y, Wei F, et al. 2016. Transcription upregulation via force-induced direct stretching of chromatin. Nat Mater, 15: 1287-1296. doi: 10.1038/nmat4729 [237] Taylor R, Hamid F, Fielding T, et al. 2022. Prematurely terminated intron-retaining mRNAs invade axons in SFPQ null-driven neurodegeneration and are a hallmark of ALS. Nat Commun, 13: 6994. doi: 10.1038/s41467-022-34331-4 [238] Thorsen T, Maerkl S J, Quake S R. 2002. Microfluidic large-scale integration. Science, 298 : 580-584. [239] Toda T, Hsu J Y, Linker S B, et al. 2017. Nup153 Interacts with Sox2 to Enable Bimodal Gene Regulation and Maintenance of Neural Progenitor Cells. Cell Stem Cell, 21 : 618-634. e617. [240] Towbin B D, González-Aguilera C, Sack R, et al. 2012. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell, 150: 934-947. doi: 10.1016/j.cell.2012.06.051 [241] Tusamda Wakhloo N, Anders S, Badique F, et al. 2020. Actomyosin, vimentin and LINC complex pull on osteosarcoma nuclei to deform on micropillar topography. Biomaterials, 234: 119746. doi: 10.1016/j.biomaterials.2019.119746 [242] Urbanska M, Muñoz H E, Shaw Bagnall J, et al. 2020. A comparison of microfluidic methods for high-throughput cell deformability measurements. Nat Methods, 17: 587-593. doi: 10.1038/s41592-020-0818-8 [243] Vahabikashi A, Adam S A, Medalia O, et al. 2022a. Nuclear lamins: Structure and function in mechanobiology. APL Bioeng, 6: 011503. doi: 10.1063/5.0082656 [244] Vahabikashi A, Sivagurunathan S, Nicdao F A S, et al. 2022b. Nuclear lamin isoforms differentially contribute to LINC complex-dependent nucleocytoskeletal coupling and whole-cell mechanics. Proc Natl Acad Sci U S A, 119: e2121816119. doi: 10.1073/pnas.2121816119 [245] Van Loenhout M T J, van der Heijden T, Kanaar R, et al. 2009. Dynamics of RecA filaments on single-stranded DNA. Nucleic Acids Research, 37: 4089-4099. doi: 10.1093/nar/gkp326 [246] Van Steensel B, Belmont A S. 2017. Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell, 169 : 780-791. [247] Vargas-Pinto R, Gong H, Vahabikashi A, et al. 2013. The effect of the endothelial cell cortex on atomic force microscopy measurements. Biophysical Journal, 105: 300-309. doi: 10.1016/j.bpj.2013.05.034 [248] Vaziri A, Mofrad M R K. 2007. Mechanics and deformation of the nucleus in micropipette aspiration experiment. Journal of Biomechanics, 40 : 2053-2062. [249] Venturini V, Pezzano F, Català Castro F, et al. 2020. The nucleus measures shape changes for cellular proprioception to control dynamic cell behavior. Science, 370: eaba2644. doi: 10.1126/science.aba2644 [250] Vial A, Costa L, Dosset P, et al. 2023. Structure and mechanics of the human nuclear pore complex basket using correlative AFM-fluorescence superresolution microscopy. Nanoscale, 15: 5756-5770. doi: 10.1039/D2NR06034E [251] Vicidomini G, Bianchini P, Diaspro A. 2018. STED super-resolved microscopy. Nat Methods, 15 : 173-182. [252] Walker C J, Crocini C, Ramirez D, et al. 2021. Nuclear mechanosensing drives chromatin remodelling in persistently activated fibroblasts. Nature Biomedical Engineering, 5: 1485-1499. doi: 10.1038/s41551-021-00709-w [253] Wang N, Tytell J D, Ingber D E. 2009. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol, 10 : 75-82. [254] Wang Y-Z, Liu C, Zhao J, et al. 2021. H2A mono-ubiquitination differentiates FACT’s functions in nucleosome assembly and disassembly. Nucleic Acids Research, 50: 833-846. [255] Wang Y, Liu M, Zhang Y, et al. 2023. Recent methods of droplet microfluidics and their applications in spheroids and organoids. Lab Chip, 23: 1080-1096. doi: 10.1039/D2LC00493C [256] Wang Y H, Hariharan A, Bastianello G, et al. 2017. DNA damage causes rapid accumulation of phosphoinositides for ATR signaling. Nat Commun, 8: 2118. doi: 10.1038/s41467-017-01805-9 [257] Waugh R, Evans E A. 1979. Thermoelasticity of red blood cell membrane. Biophys J, 26 : 115-131. [258] Weber I P, Yun S H, Scarcelli G, et al. 2017. The role of cell body density in ruminant retina mechanics assessed by atomic force and Brillouin microscopy. Phys Biol, 14: 065006. doi: 10.1088/1478-3975/aa6d18 [259] Wei F, Xu X, Zhang C, et al. 2020a. Stress fiber anisotropy contributes to force-mode dependent chromatin stretching and gene upregulation in living cells. Nature Communications, 11: 4902. doi: 10.1038/s41467-020-18584-5 [260] Wei M, Fan X, Ding M, et al. 2020b. Nuclear actin regulates inducible transcription by enhancing RNA polymerase II clustering. Sci Adv, 6: eaay6515. doi: 10.1126/sciadv.aay6515 [261] Werdich A A, Lima E A, Ivanov B, et al. 2004. A microfluidic device to confine a single cardiac myocyte in a sub-nanoliter volume on planar microelectrodes for extracellular potential recordings. Lab Chip, 4: 357-362. doi: 10.1039/b315648f [262] Wintner O, Hirsch-Attas N, Schlossberg M, et al. 2020. A unified linear viscoelastic model of the Ccell nucleus defines the mechanical contributions of lamins and chromatin. Adv Sci (Weinh), 7: 1901222. doi: 10.1002/advs.201901222 [263] Wijesinghe W C B, Min D. 2023. Single-Molecule Force Spectroscopy of Membrane Protein Folding. J Mol Biol, 435: 167975 [264] Woerner A C, Frottin F, Hornburg D, et al. 2016. Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science, 351: 173-176. doi: 10.1126/science.aad2033 [265] Wong X, Stewart C L. 2020. The laminopathies and the insights they provide into the structural and functional organization of the nucleus. Annu Rev Genomics Hum Genet, 21 : 263-288. [266] Wu P-H, Aroush D R-B, Asnacios A, et al. 2018. A comparison of methods to assess cell mechanical properties. Nature Methods, 15: 491-498. doi: 10.1038/s41592-018-0015-1 [267] Wu Y, Shroff H. 2018. Faster, sharper, and deeper: structured illumination microscopy for biological imaging. Nat Methods, 15 : 1011-1019. [268] Xu J, Tseng Y, Wirtz D. 2000. Strain hardening of actin filament networks: Regulation by the dynamic cross-linking protein α-ACTININ. Journal of Biological Chemistry, 275 : 35886-35892. [269] Yamada S, Wirtz D, Coulombe P A. 2003. The mechanical properties of simple epithelial keratins 8 and 18: discriminating between interfacial and bulk elasticities. Journal of Structural Biology, 143 : 45-55. [270] Yang C, Tibbitt M W, Basta L, et al. 2014. Mechanical memory and dosing influence stem cell fate. Nat Mater, 13: 645-652. doi: 10.1038/nmat3889 [271] Yang F, Bevilacqua C, Hambura S, et al. 2023. Pulsed stimulated Brillouin microscopy enables high-sensitivity mechanical imaging of live and fragile biological specimens. Nature Methods. [272] Yue X, Cui J, Sun Z, et al. 2023. Nuclear softening mediated by Sun2 suppression delays mechanical stress-induced cellular senescence. Cell Death Discov, 9: 167. doi: 10.1038/s41420-023-01467-1 [273] Yusuf M, Kaneyoshi K, Fukui K, et al. 2019. Use of 3D imaging for providing insights into high-order structure of mitotic chromosomes. Chromosoma, 128: 7-13. doi: 10.1007/s00412-018-0678-5 [274] Zhang J, Nou X A, Kim H, et al. 2017. Brillouin flow cytometry for label-free mechanical phenotyping of the nucleus. Lab Chip, 17: 663-670. doi: 10.1039/C6LC01443G [275] Zhang J, Raghunathan R, Rippy J, et al. 2019. Tissue biomechanics during cranial neural tube closure measured by Brillouin microscopy and optical coherence tomography. Birth Defects Res, 111: 991-998. doi: 10.1002/bdr2.1389 [276] Zhang W, Wang Y, Liu Y, et al. 2023. NFIB facilitates replication licensing by acting as a genome organizer. Nature Communications, 14: 5076. doi: 10.1038/s41467-023-40846-1 [277] Zhang X, Chen S, Yoo S, et al. 2008. Mutation in nuclear pore component NUP155 leads to atrial fibrillation and early sudden cardiac death. Cell, 135: 1017-1027. doi: 10.1016/j.cell.2008.10.022 [278] Zheng M, Tian S Z, Capurso D, et al. 2019. Multiplex chromatin interactions with single-molecule precision. Nature, 566: 558-562. doi: 10.1038/s41586-019-0949-1 [279] Zhou L, Panté N. 2010. The nucleoporin Nup153 maintains nuclear envelope architecture and is required for cell migration in tumor cells. FEBS Lett, 584 : 3013-3020. [280] Zhou Z L, Hui T H, Tang B, et al. 2014. Accurate measurement of stiffness of leukemia cells and leukocytes using an optical trap by a rate-jump method. RSC Advances, 4: 8453-8460. doi: 10.1039/c3ra45835k [281] Zhu X, Huang G, Zeng C, et al. 2022. Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex. Science, 376: eabl8280. doi: 10.1126/science.abl8280 [282] Zidovska A, Weitz D A, Mitchison T J. 2013. Micron-scale coherence in interphase chromatin dynamics. Proc Natl Acad Sci U S A, 110: 15555-15560. doi: 10.1073/pnas.1220313110 [283] Zimmerli C E, Allegretti M, Rantos V, et al. 2021. Nuclear pores dilate and constrict in cellulo. Science, 374: eabd9776. doi: 10.1126/science.abd9776 [284] Zwerger M, Ho C Y, Lammerding J. 2011. Nuclear mechanics in disease. Annual Review of Biomedical Engineering, 13 : 397-428. -



 下载:
下载: